Table of Links
-
Materials and Methods
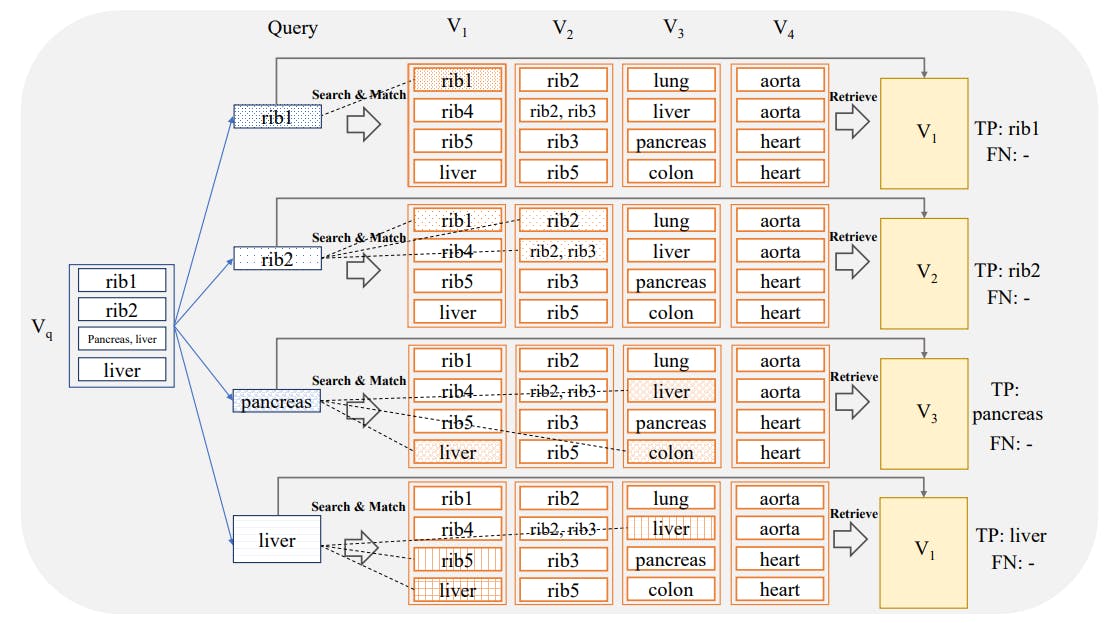
2.1 Vector Database and Indexing
-
Discussion
4.1 Dataset and 4.2 Re-ranking
4.4 Volume-based, Region-based and Localized Retrieval and 4.5 Localization-ratio
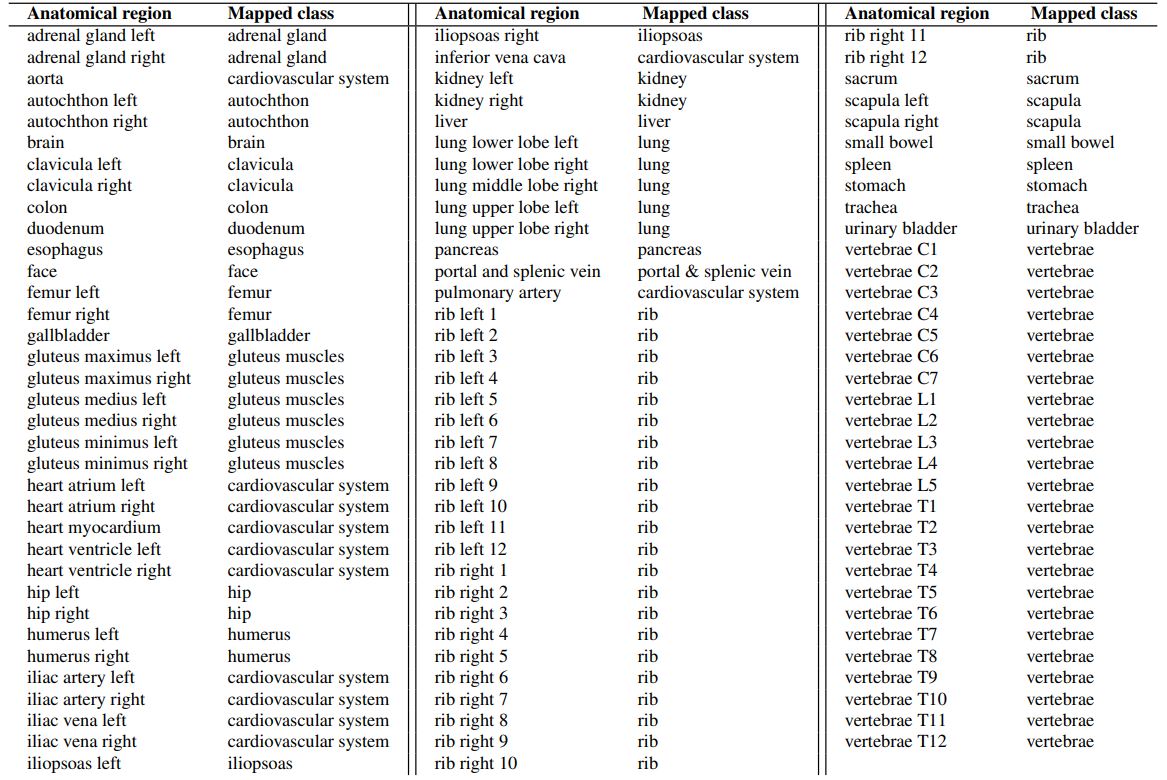
2.2 Feature Extractors
We extend the analysis of Khun Jush et al. [2023] by adding two ResNet50 embeddings and evaluating the performance of six different slice embedding extractors for CBIR tasks. All the feature extractors are based on deep-learning-based models.
Self-supervised Models: We employed three self-supervised models pre-trained on ImageNet [Deng et al., 2009]. DINOv1 [Caron et al., 2021], that demonstrated learning efficient image representations from unlabeled data using self-distillation. DINOv2 [Oquab et al., 2023], is built upon DINOv1 [Caron et al., 2021], and this model scales the pre-training process by combining an improved training dataset, patchwise objectives during training and introducing a new regularization technique, which gives rise to superior performance on segmentation tasks. DreamSim [Fu et al., 2023], built upon the foundation of DINOv1 [Caron et al., 2021], fine-tunes the model using synthetic data triplets specifically designed to be cognitively impenetrable with human judgments. For the self-supervised models, we used the best-performing backbone reported by the developers of the models.
Supervised Models: We included a SwinTransformer model [Liu et al., 2021] and a ResNet50 model [He et al., 2016] trained in a supervised manner using the RadImageNet dataset [Mei et al., 2022] that includes 5 million annotated 2D CT, MRI, and ultrasound images of musculoskeletal, neurologic, oncologic, gastrointestinal, endocrine, and pulmonary pathology. Furthermore, a ResNet50 model pre-trained on rendered images of fractal geometries was included based on [Kataoka et al., 2022]. These training images are formula-derived, non-natural, and do not require any human annotation.
Authors:
(1) Farnaz Khun Jush, Bayer AG, Berlin, Germany ([email protected]);
(2) Steffen Vogler, Bayer AG, Berlin, Germany ([email protected]);
(3) Tuan Truong, Bayer AG, Berlin, Germany ([email protected]);
(4) Matthias Lenga, Bayer AG, Berlin, Germany ([email protected]).
This paper is