Table of Links
2 MindEye2 and 2.1 Shared-Subject Functional Alignment
2.2 Backbone, Diffusion Prior, & Submodules
2.3 Image Captioning and 2.4 Fine-tuning Stable Diffusion XL for unCLIP
3 Results and 3.1 fMRI-to-Image Reconstruction
3.3 Image/Brain Retrieval and 3.4 Brain Correlation
6 Acknowledgements and References
A Appendix
A.2 Additional Dataset Information
A.3 MindEye2 (not pretrained) vs. MindEye1
A.4 Reconstruction Evaluations Across Varying Amounts of Training Data
A.5 Single-Subject Evaluations
A.7 OpenCLIP BigG to CLIP L Conversion
A.9 Reconstruction Evaluations: Additional Information
A.10 Pretraining with Less Subjects
A.11 UMAP Dimensionality Reduction
A.13 Human Preference Experiments
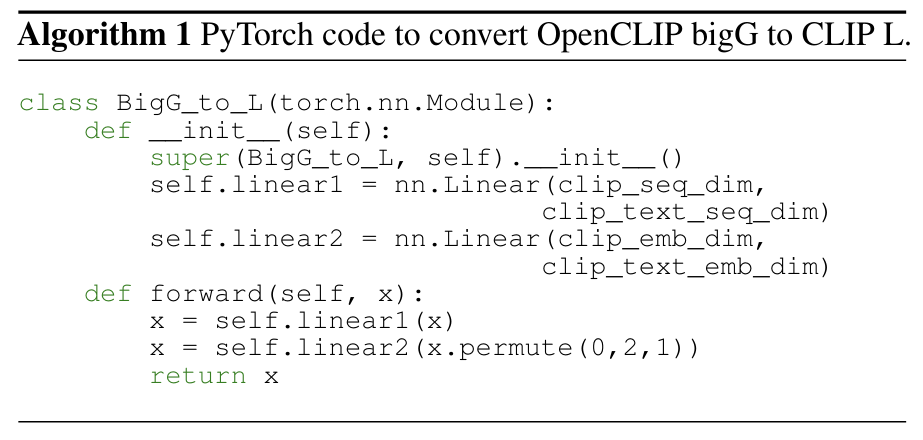
A.7 OpenCLIP BigG to CLIP L Conversion
To map from OpenCLIP ViT-bigG/14 image latents to CLIP ViT-L/14 image latents during MindEye2 inference we inde


pendently trained a linear model using ground truth images from the COCO 2017 train and validation dataset. This conversion was necessary to use the pretrained GIT image captioning model. The PyTorch code used to train this model is depicted in Algorithm 1.
This paper is available on arxiv under CC BY 4.0 DEED license.
Authors:
(1) Paul S. Scotti, Stability AI and Medical AI Research Center (MedARC);
(2) Mihir Tripathy, Medical AI Research Center (MedARC) and a Core contribution;
(3) Cesar Kadir Torrico Villanueva, Medical AI Research Center (MedARC) and a Core contribution;
(4) Reese Kneeland, University of Minnesota and a Core contribution;
(5) Tong Chen, The University of Sydney and Medical AI Research Center (MedARC);
(6) Ashutosh Narang, Medical AI Research Center (MedARC);
(7) Charan Santhirasegaran, Medical AI Research Center (MedARC);
(8) Jonathan Xu, University of Waterloo and Medical AI Research Center (MedARC);
(9) Thomas Naselaris, University of Minnesota;
(10) Kenneth A. Norman, Princeton Neuroscience Institute;
(11) Tanishq Mathew Abraham, Stability AI and Medical AI Research Center (MedARC).
